Taking Data Apps into WebApp: Using Streamlit, Plotly, and Python
Introduction
From the past 2 stories of a data and its journey to confess the insights, we have explored several areas and to point out few:
- We have done EDA based on descriptive and inferential part of the statistics to find strong evidences, relationships and facts about the data.
- We used some of valuable insights from the EDA and tried to classify the possible environment that the properties reflects to. One example is, we tried to predict the value of CO based on Smoke and LPG.
But now in this part, we will try to take those experiments into web app where we could tweak different aspects our experiment by making a simple yet powerful web app using Streamlit. Streamlit is a free tool available in Python that allows us to make Data Apps faster.
Making Things Ready
- Please install Streamlit by doing
pip install streamlit. - Once installed, please make sure it is recognized by system as a environment variable by doing
streamlit --versionand if it gives a output, then we are ready to go. - Please install Plotly as we will be making interactive plots based on it.
Getting Data Ready
For this purpose, we will be working with Room Occupancy Detection Data. Which is similar to the previous data. There are 3 text files with CSV formats, datatraining.txt, datatest.txt and datatest2.txt. Lets read them using Pandas and convert the date column to datetime. The column Occupancy contains a binary value 0/1 which will be the label for us later on.
import pandas as pd
import cufflinks
import plotly.io as pio
import warnings
import numpy as np
cufflinks.go_offline()
cufflinks.set_config_file(world_readable=True, theme='pearl')
pio.renderers.default = "notebook"
warnings.simplefilter("ignore")
train = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatraining.txt")
train["date"]=pd.to_datetime(train.date)
train
| date | Temperature | Humidity | Light | CO2 | HumidityRatio | Occupancy | |
|---|---|---|---|---|---|---|---|
| 1 | 2015-02-04 17:51:00 | 23.18 | 27.2720 | 426.0 | 721.250000 | 0.004793 | 1 |
| 2 | 2015-02-04 17:51:59 | 23.15 | 27.2675 | 429.5 | 714.000000 | 0.004783 | 1 |
| 3 | 2015-02-04 17:53:00 | 23.15 | 27.2450 | 426.0 | 713.500000 | 0.004779 | 1 |
| 4 | 2015-02-04 17:54:00 | 23.15 | 27.2000 | 426.0 | 708.250000 | 0.004772 | 1 |
| 5 | 2015-02-04 17:55:00 | 23.10 | 27.2000 | 426.0 | 704.500000 | 0.004757 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 8139 | 2015-02-10 09:29:00 | 21.05 | 36.0975 | 433.0 | 787.250000 | 0.005579 | 1 |
| 8140 | 2015-02-10 09:29:59 | 21.05 | 35.9950 | 433.0 | 789.500000 | 0.005563 | 1 |
| 8141 | 2015-02-10 09:30:59 | 21.10 | 36.0950 | 433.0 | 798.500000 | 0.005596 | 1 |
| 8142 | 2015-02-10 09:32:00 | 21.10 | 36.2600 | 433.0 | 820.333333 | 0.005621 | 1 |
| 8143 | 2015-02-10 09:33:00 | 21.10 | 36.2000 | 447.0 | 821.000000 | 0.005612 | 1 |
test1 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest.txt")
test1["date"]=pd.to_datetime(test1.date)
test1
| date | Temperature | Humidity | Light | CO2 | HumidityRatio | Occupancy | |
|---|---|---|---|---|---|---|---|
| 140 | 2015-02-02 14:19:00 | 23.700000 | 26.272000 | 585.200000 | 749.200000 | 0.004764 | 1 |
| 141 | 2015-02-02 14:19:59 | 23.718000 | 26.290000 | 578.400000 | 760.400000 | 0.004773 | 1 |
| 142 | 2015-02-02 14:21:00 | 23.730000 | 26.230000 | 572.666667 | 769.666667 | 0.004765 | 1 |
| 143 | 2015-02-02 14:22:00 | 23.722500 | 26.125000 | 493.750000 | 774.750000 | 0.004744 | 1 |
| 144 | 2015-02-02 14:23:00 | 23.754000 | 26.200000 | 488.600000 | 779.000000 | 0.004767 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 2800 | 2015-02-04 10:38:59 | 24.290000 | 25.700000 | 808.000000 | 1150.250000 | 0.004829 | 1 |
| 2801 | 2015-02-04 10:40:00 | 24.330000 | 25.736000 | 809.800000 | 1129.200000 | 0.004848 | 1 |
| 2802 | 2015-02-04 10:40:59 | 24.330000 | 25.700000 | 817.000000 | 1125.800000 | 0.004841 | 1 |
| 2803 | 2015-02-04 10:41:59 | 24.356667 | 25.700000 | 813.000000 | 1123.000000 | 0.004849 | 1 |
| 2804 | 2015-02-04 10:43:00 | 24.408333 | 25.681667 | 798.000000 | 1124.000000 | 0.004860 | 1 |
test2 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest2.txt")
test2["date"]=pd.to_datetime(test2.date)
test2
| date | Temperature | Humidity | Light | CO2 | HumidityRatio | Occupancy | |
|---|---|---|---|---|---|---|---|
| 1 | 2015-02-11 14:48:00 | 21.7600 | 31.133333 | 437.333333 | 1029.666667 | 0.005021 | 1 |
| 2 | 2015-02-11 14:49:00 | 21.7900 | 31.000000 | 437.333333 | 1000.000000 | 0.005009 | 1 |
| 3 | 2015-02-11 14:50:00 | 21.7675 | 31.122500 | 434.000000 | 1003.750000 | 0.005022 | 1 |
| 4 | 2015-02-11 14:51:00 | 21.7675 | 31.122500 | 439.000000 | 1009.500000 | 0.005022 | 1 |
| 5 | 2015-02-11 14:51:59 | 21.7900 | 31.133333 | 437.333333 | 1005.666667 | 0.005030 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 9748 | 2015-02-18 09:15:00 | 20.8150 | 27.717500 | 429.750000 | 1505.250000 | 0.004213 | 1 |
| 9749 | 2015-02-18 09:16:00 | 20.8650 | 27.745000 | 423.500000 | 1514.500000 | 0.004230 | 1 |
| 9750 | 2015-02-18 09:16:59 | 20.8900 | 27.745000 | 423.500000 | 1521.500000 | 0.004237 | 1 |
| 9751 | 2015-02-18 09:17:59 | 20.8900 | 28.022500 | 418.750000 | 1632.000000 | 0.004279 | 1 |
| 9752 | 2015-02-18 09:19:00 | 21.0000 | 28.100000 | 409.000000 | 1864.000000 | 0.004321 | 1 |
Lets look over these data and do necessary actions.
test2.date.describe()
count 9752
unique 9752
top 2015-02-15 15:04:59
freq 1
first 2015-02-11 14:48:00
last 2015-02-18 09:19:00
Name: date, dtype: object
test1.date.describe()
count 2665
unique 2665
top 2015-02-03 14:45:59
freq 1
first 2015-02-02 14:19:00
last 2015-02-04 10:43:00
Name: date, dtype: object
train.date.describe()
count 8143
unique 8143
top 2015-02-07 20:26:59
freq 1
first 2015-02-04 17:51:00
last 2015-02-10 09:33:00
Name: date, dtype: object
Looking over the date of each dataframe, the train data have data from 04 to 10 day, and test1 have 02 to 04 then test2 have 11 to 18 day. It might be best idea to concatenate train and test2 but lets explore it later on.
Exploratory Data Analysis
Missing Values
dfs = {"train":train,"test1":test1,"test2":test2}
for df in dfs.values():
total = df.isnull().sum().sort_values(ascending = False)
percent = (df.isnull().sum()/df.isnull().count()).sort_values(ascending = False)
mdf = pd.concat([total, percent], axis=1, keys=['Total', 'Percent'])
mdf = mdf.reset_index()
print(mdf)
index Total Percent
0 date 0 0.0
1 Temperature 0 0.0
2 Humidity 0 0.0
3 Light 0 0.0
4 CO2 0 0.0
5 HumidityRatio 0 0.0
6 Occupancy 0 0.0
index Total Percent
0 date 0 0.0
1 Temperature 0 0.0
2 Humidity 0 0.0
3 Light 0 0.0
4 CO2 0 0.0
5 HumidityRatio 0 0.0
6 Occupancy 0 0.0
index Total Percent
0 date 0 0.0
1 Temperature 0 0.0
2 Humidity 0 0.0
3 Light 0 0.0
4 CO2 0 0.0
5 HumidityRatio 0 0.0
6 Occupancy 0 0.0
It seems that there is no missing values in each columns. Lets see the distribution of each columns.
Summary of Each Variables
For the purpose of comparing distribution of values in each dataframe, we will plot boxplot side by side.
Please ignore the import time and fig.write_image(..) part.
from plotly.subplots import make_subplots
import plotly.graph_objects as go
import time
titles = list(dfs.keys())
for c in train.columns:
if c!="date":
fig = make_subplots(rows=2,cols=3, subplot_titles=titles, )
fig.add_trace(go.Box(y=train[c].tolist(), name=titles[0]), row=1, col=1)
fig.add_trace(go.Box(y=test1[c].tolist(), name = titles[1]), row=1, col=2)
fig.add_trace(go.Box(y=test2[c].tolist(), name = titles[2]), row=1, col=3)
fig.add_trace(go.Histogram(y=train[c].tolist(), name=titles[0]), row=2, col=1)
fig.add_trace(go.Histogram(y=test1[c].tolist(), name = titles[1]), row=2, col=2)
fig.add_trace(go.Histogram(y=test2[c].tolist(), name = titles[2]), row=2, col=3)
fig.update_layout(height=600, width=800, title_text=f"Box and Distribution of {c}")
fig.show()
fig.write_image(f"summary_{c}.png")
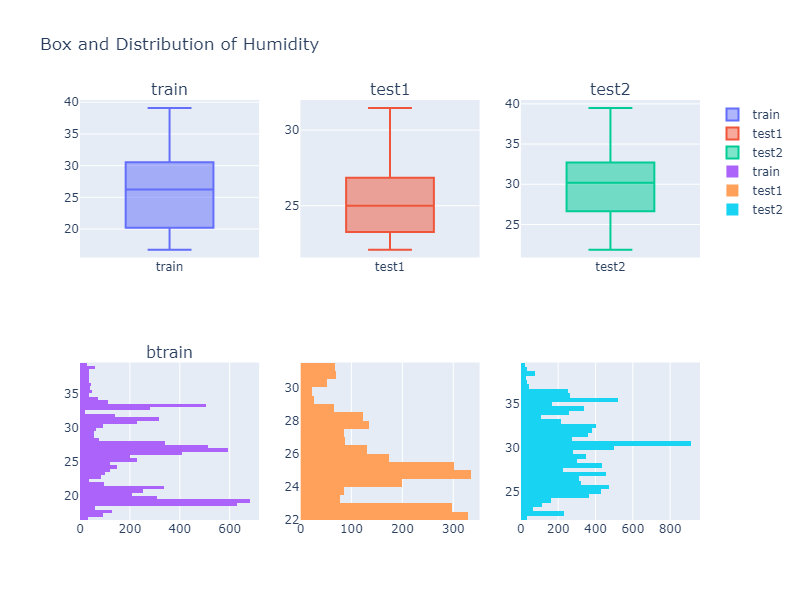
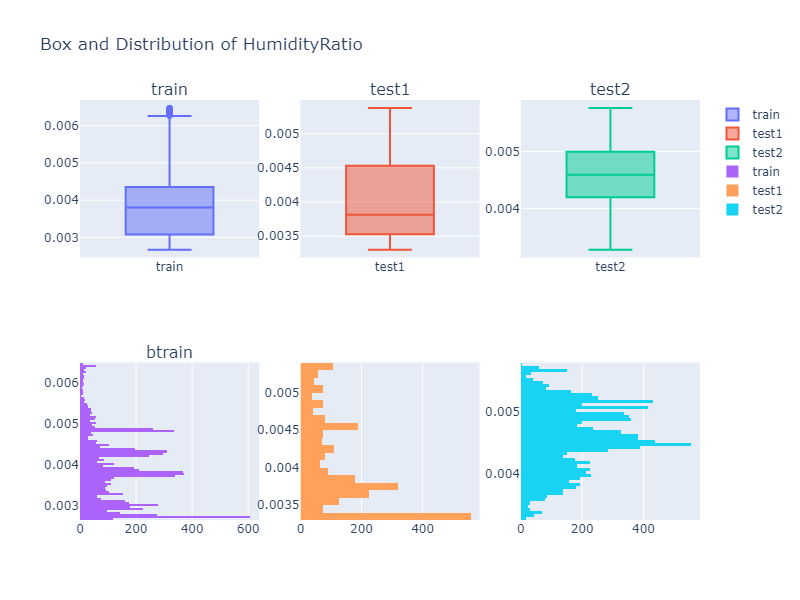
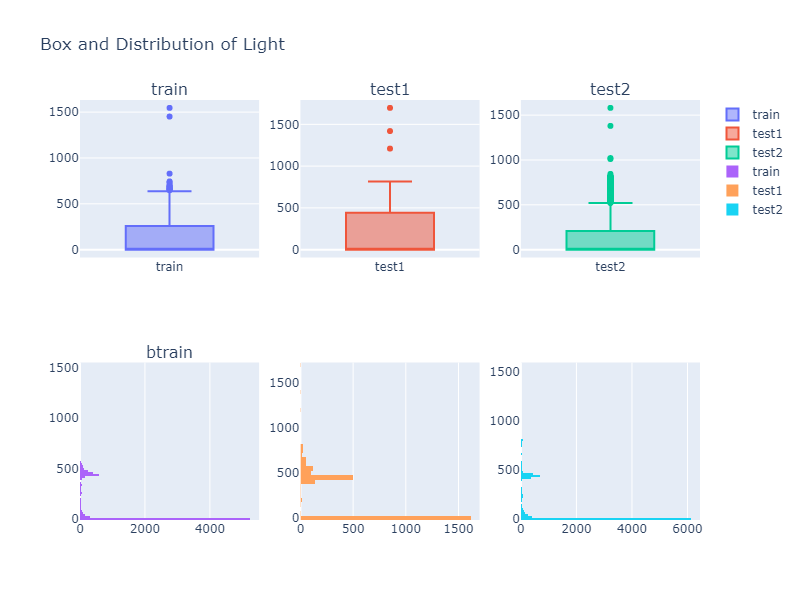
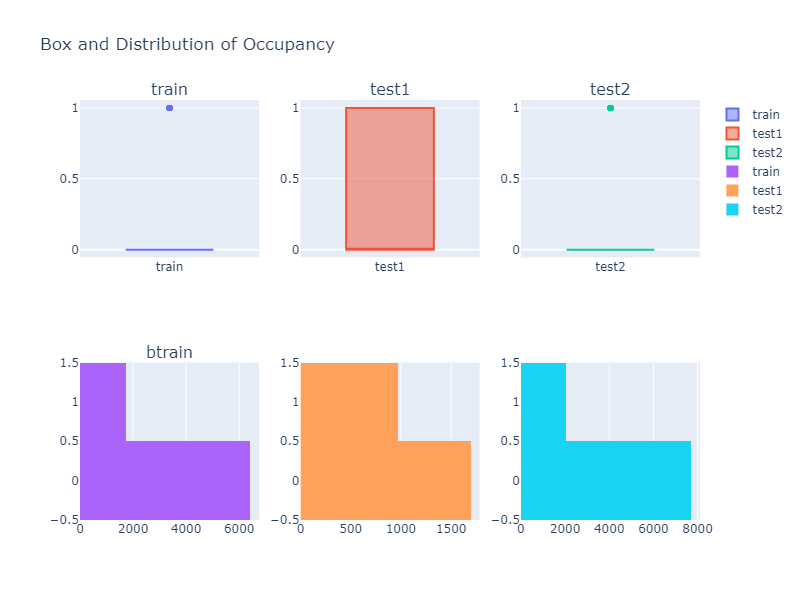
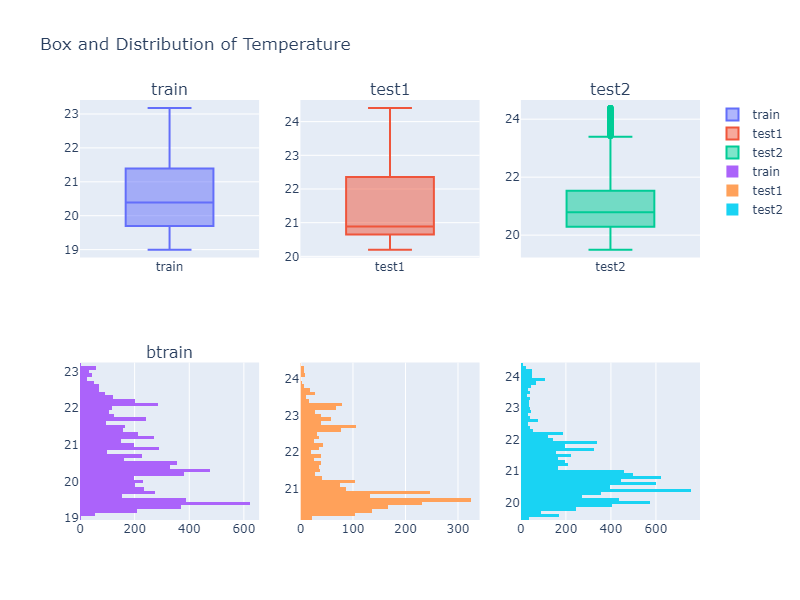
Looking over a Histogram and a box plot of different column values, we can see that the descriptive property of a data is not identical to each other. Thus we might need to do some kind of data transformation if our model does not perform well.
Correlation
Lets see if correlation between variables are same and if they do, we will be on the bright side.
fig = make_subplots(rows=1,cols=3, subplot_titles=titles)
fig.add_trace(go.Heatmap(z=train.corr(), y=train.corr().columns,x=train.corr().index, name=titles[0]), row=1, col=1)
fig.add_trace(go.Heatmap(z=test1.corr(), x=train.corr().index, name = titles[1]), row=1, col=2)
fig.add_trace(go.Heatmap(z=test2.corr(), x=train.corr().index, name = titles[2]), row=1, col=3)
fig.show()
fig.write_image(f"corr.png")
The correlation seems almost similar for all 3 dataframes.
Taking EDA to Streamlit App
Please create a project folder and inside it, create a Python file main.py. This file will be our main file where we will do all these plots and it will take our plots, analysis into the web app.
First Streamlit App
We will read our file and then put it in a cache so that we wont have to read it whenever our app is changed.
import streamlit as st
import numpy as np
import pandas as pd
import cufflinks
@st.cache
def get_data():
train = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatraining.txt")
train["date"]=pd.to_datetime(train.date)
test1 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest.txt")
test1["date"]=pd.to_datetime(test1.date)
test2 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest2.txt")
test2["date"]=pd.to_datetime(test2.date)
dfs = {"train":train,"test1":test1,"test2":test2}
return dfs
dfs = get_data()
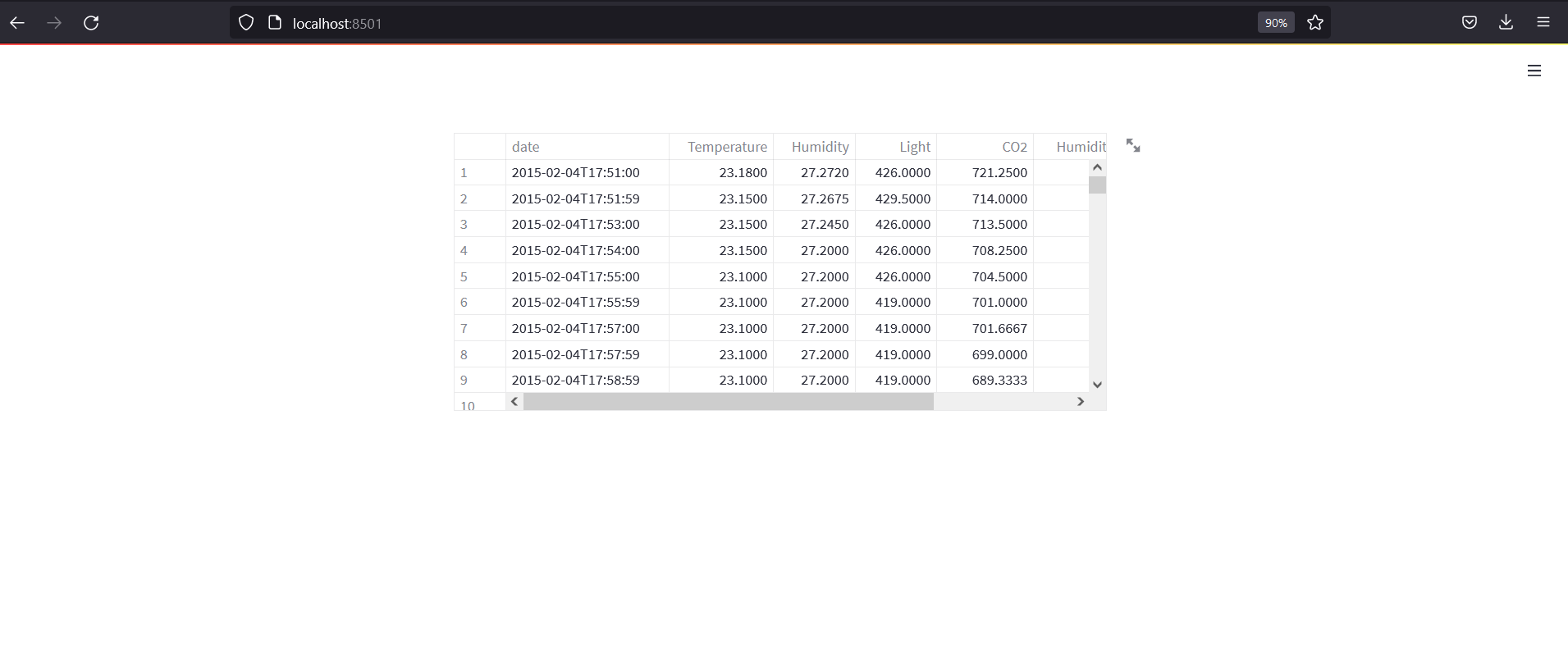
st.dataframe(dfs["train"])
In above code, we have read 3 files and put them in dictionary as dfs the returned. The @st.cache decorator allows us to cache the file so that we wont need to reload the data whenever the app reloads. Then we have shown the dataframe in a app. App should look like below:
For the next step, we will add few select box and then the analysis parts.
import streamlit as st
import numpy as np
import pandas as pd
import cufflinks
from plotly.subplots import make_subplots
import plotly.graph_objects as go
@st.cache
def get_data():
train = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatraining.txt")
train["date"]=pd.to_datetime(train.date)
test1 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest.txt")
test1["date"]=pd.to_datetime(test1.date)
test2 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest2.txt")
test2["date"]=pd.to_datetime(test2.date)
dfs = {"train":train,"test1":test1,"test2":test2}
return dfs
dfs = get_data()
sidebar = st.sidebar
# select modes, EDA, Clustering, Regression and Classification
mode = sidebar.selectbox("Select a mode.",options=["EDA", "Clustering", "Regression", "Classification"])
# If selected EDA, show EDA related plots
if mode=="EDA":
# if selected show the data
show_data = sidebar.checkbox("Show data")
if show_data:
# if selected, show train data
if sidebar.checkbox("Show Train data"):
st.markdown("### Train Data")
st.dataframe(dfs["train"])
# if selected, show test1 data
if sidebar.checkbox("Show Test1 data"):
st.markdown("### Test1 Data")
st.dataframe(dfs["test1"])
# if selected, show test2 data
if sidebar.checkbox("Show Test2 data"):
st.markdown("### Test2 Data")
st.dataframe(dfs["test2"])
# if selected, show the comparision data
show_comparison = sidebar.checkbox("Show comparison")
if show_comparison:
# make a multiselect to select the columns to compare
selected = sidebar.multiselect("Select Columns ", [d for d in dfs["train"].columns if d not in ["date"]])
titles=list(dfs.keys())
train = dfs["train"]
test1 = dfs["test1"]
test2 = dfs["test2"]
if selected:
st.markdown(f"### Selected Columns: {', '.join(selected)}")
for c in selected:
fig = make_subplots(rows=2,cols=3, subplot_titles=titles, )
fig.add_trace(go.Box(y=train[c].tolist(), name=titles[0]), row=1, col=1)
fig.add_trace(go.Box(y=test1[c].tolist(), name = titles[1]), row=1, col=2)
fig.add_trace(go.Box(y=test2[c].tolist(), name = titles[2]), row=1, col=3)
fig.add_trace(go.Histogram(y=train[c].tolist(), name=titles[0]), row=2, col=1)
fig.add_trace(go.Histogram(y=test1[c].tolist(), name = titles[1]), row=2, col=2)
fig.add_trace(go.Histogram(y=test2[c].tolist(), name = titles[2]), row=2, col=3)
fig.update_layout(height=600, width=800, title_text=f"Box and Distribution of {c}")
st.plotly_chart(fig)
# if selected show correlation
show_corr = sidebar.checkbox("Show Correlation")
if show_corr:
st.markdown("### Correlation")
fig = make_subplots(rows=1,cols=3, subplot_titles=titles)
fig.add_trace(go.Heatmap(z=train.corr(), y=train.corr().columns,x=train.corr().index, name=titles[0]), row=1, col=1)
fig.add_trace(go.Heatmap(z=test1.corr(), x=train.corr().index, name = titles[1]), row=1, col=2)
fig.add_trace(go.Heatmap(z=test2.corr(), x=train.corr().index, name = titles[2]), row=1, col=3)
st.plotly_chart(fig)
In above code, we have added everything we did on EDA into a web app. We have added a comment above the part of code that needs explanation. Now our app looks like below:
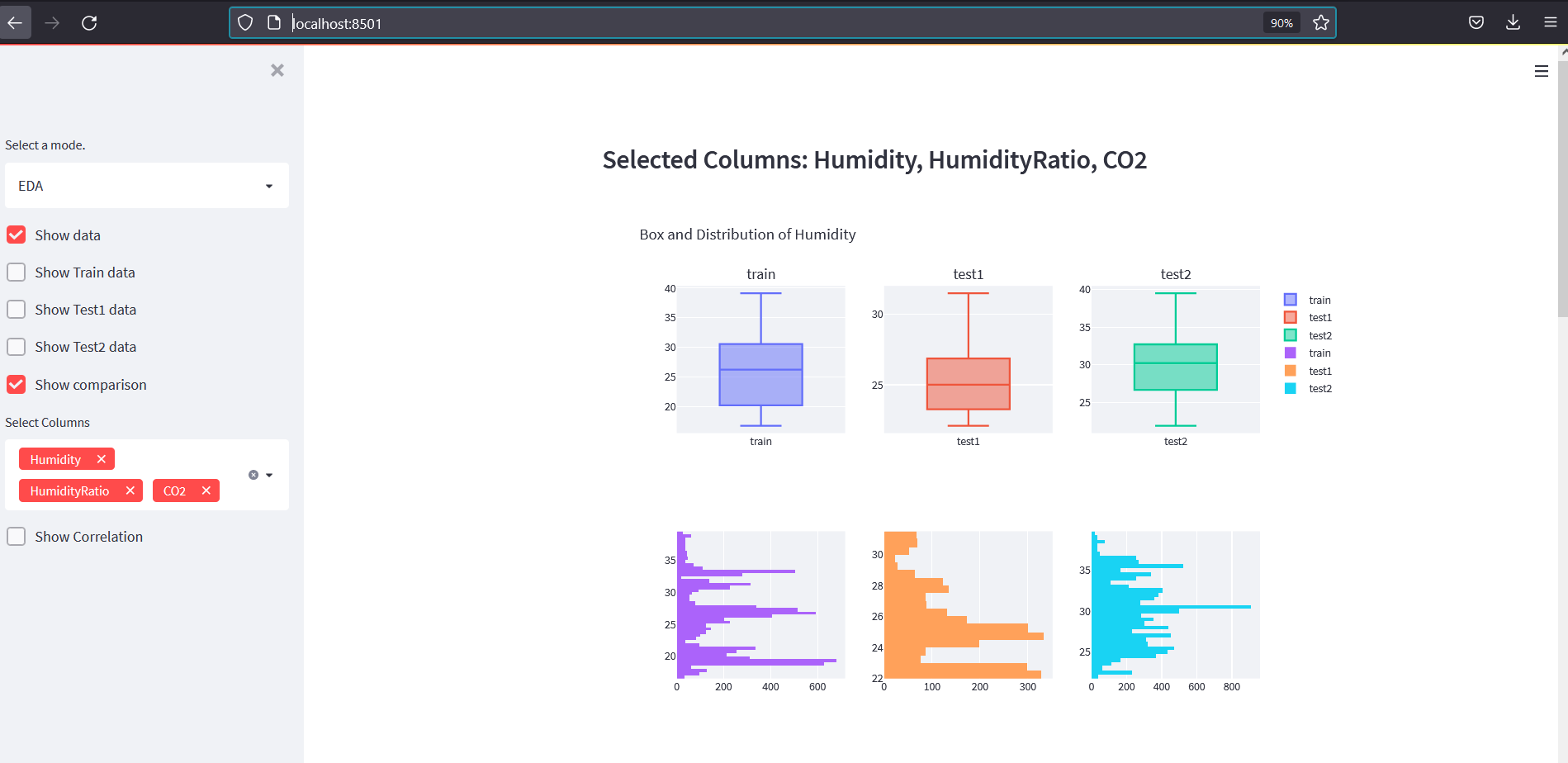
Clustering
Now we want to cluster our data based on the features we have. We already know that there are two classes in data occupancy, but lets try to find if some kind of clusters can be seen or not.
KMeans Clustering
Lets first do clustering based on default features and see the performance of it on the train dataframe.
from sklearn.cluster import KMeans
clusters = 5
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"][:-2]
inertias = []
for c in range(2,clusters+1):
tdf = train.copy()
X = tdf[features].to_numpy()
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=tdf[features[0]],y=tdf[features[1]],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
fig.show()
fig.write_image(f"kmeans_{c}.png")
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
fig.show()
fig.write_image(f"kmeans_cvi{c}.png")
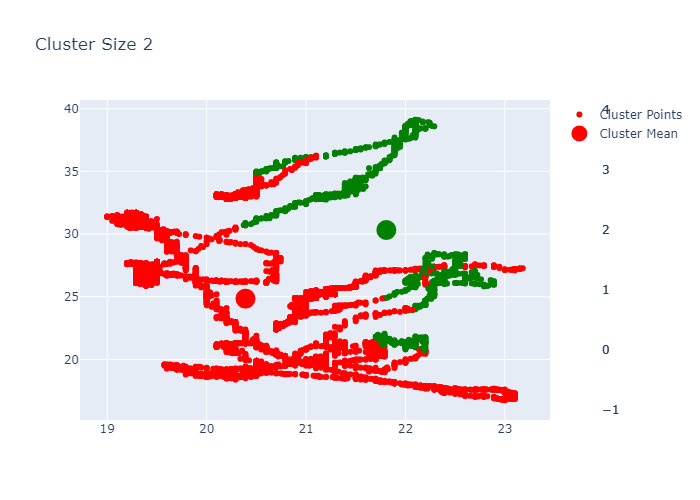
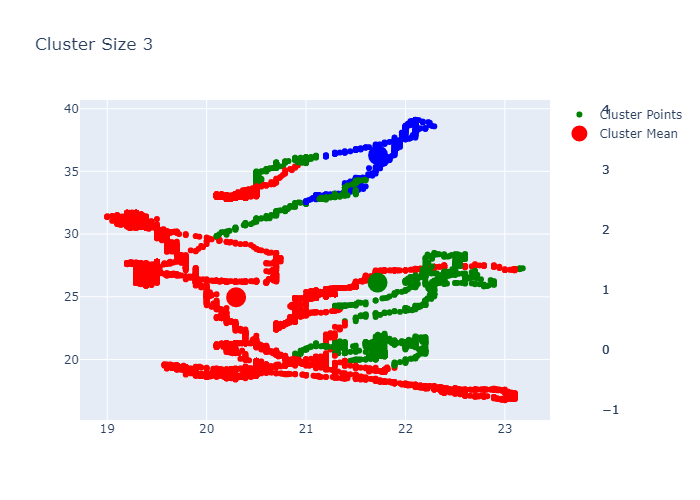
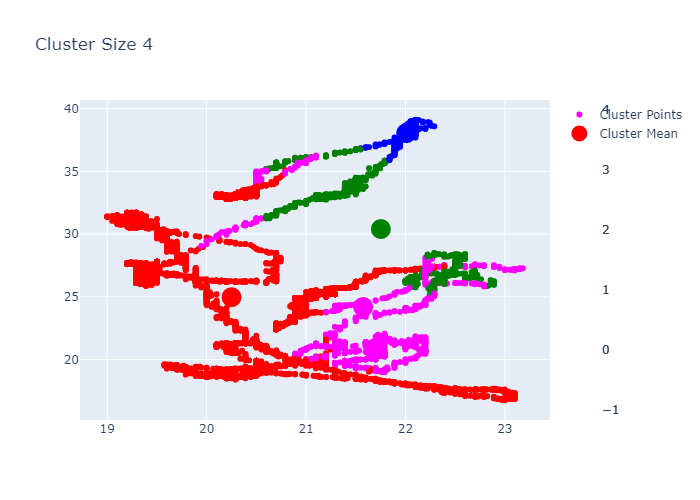
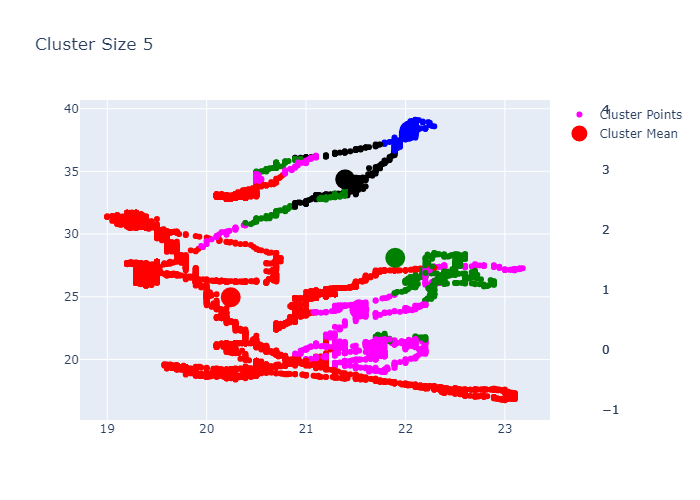
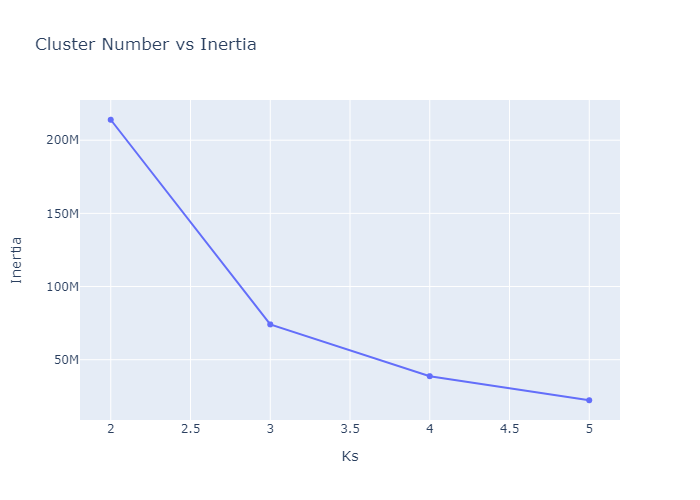
Looking over the Inertia plot, it seems that inertia has decreased slowly from ks 3. But We already know that data is from two different occupancy. The cluster plots does not seems to be great because we have multiple features used for clustering and plot is 2d. Now lets try to do dimension reduction and see the performance.
PCA for Dimensionality Reduction
PCA is used to reduce the high dimension of the data into more robust features.
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
X = train[features]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
# Create a PCA instance: pca
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)# Plot the explained variances
feat = range(pca.n_components_)
plt.bar(feat, pca.explained_variance_ratio_, color='black')
plt.xlabel('PCA features')
plt.ylabel('variance %')
plt.xticks(feat)# Save components to a DataFrame
PCA_components = pd.DataFrame(principalComponents, columns=list(range(len(feat))))
plt.show()
plt.scatter(PCA_components[1], PCA_components[2], alpha=.1, color='black')
plt.xlabel('PCA 1')
plt.ylabel('PCA 2')

Text(0, 0.5, 'PCA 2')

Looking over the plots of components, we can see some kind of clustering. Thus, we will try to make a Cluster now.
clusters = 5
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"][:-2]
inertias = []
for c in range(2,clusters+1):
X = PCA_components[[1,2]]
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
fig.show()
fig.write_image(f"pca_kmeans_{c}1.png")
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
fig.show()
fig.write_image(f"pca_kmeans_cvi.png")




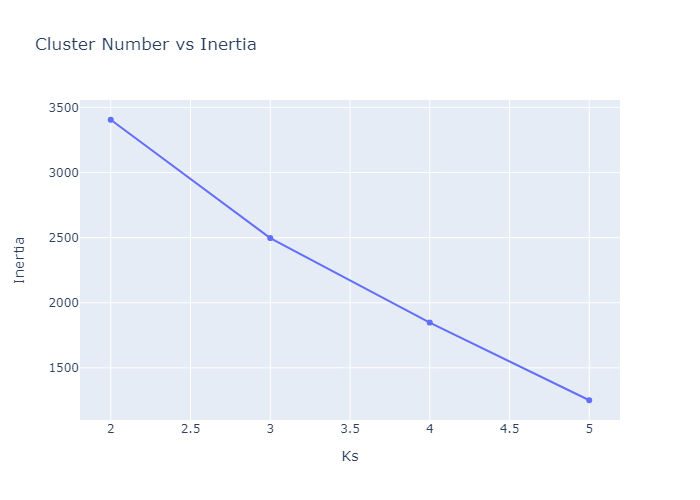
Now we can see the performance in better way. We can make cluster of 2. Before adding this into the web app, lets do KMedoids first.
KMedoids Clustering
We have already covered the theory on previous part but in this one, we will just import KMedoids from sklearn_extra and use it just like the previous part.
from sklearn_extra.cluster import KMedoids
clusters = 5
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"][:-2]
inertias = []
for c in range(2,clusters+1):
X = PCA_components[[1,2]]
colors=['red','green','blue','magenta','black','yellow']
model = KMedoids(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
fig.show()
fig.write_image(f"kmedoids_{c}.png")
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
fig.show()
fig.write_image(f"kmedoids_kvi.png")
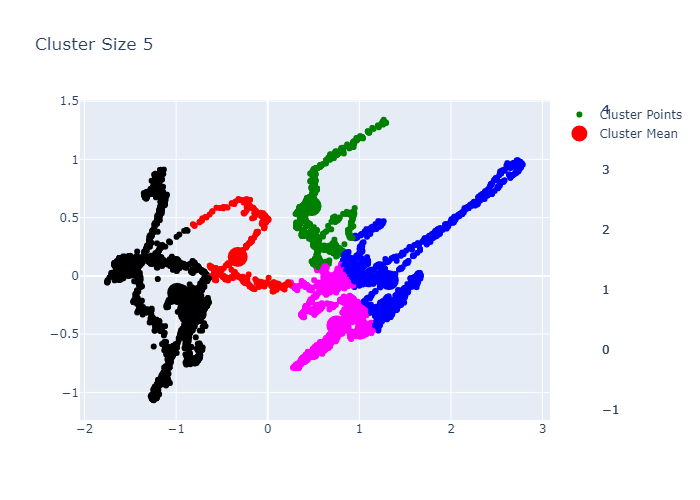
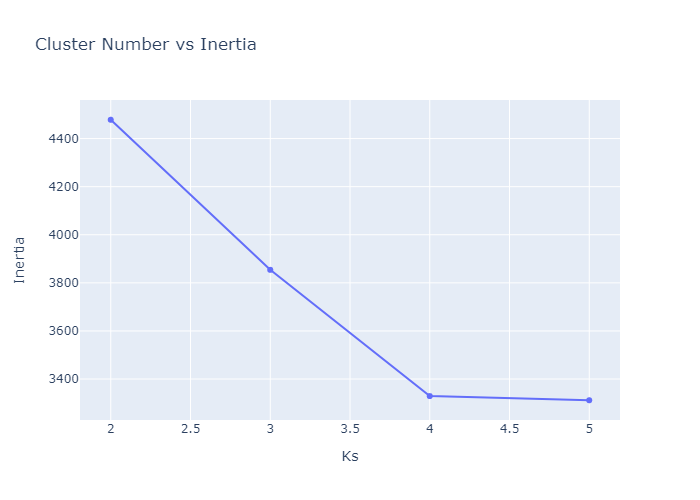
Lets add this into streamlit app now.
Taking Clustering to Streamlit App
Since we have already made a selectbox of each mode, we will add entire clustering codes in a clustering.
from sklearn_extra.cluster import KMedoids
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
if mode=="Clustering":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"][:-2]
st.markdown("## Clustering Mode Selected")
st.markdown(hline)
# select a clustering algorithm
calg = sidebar.selectbox("Select a clustering algorithm", ["K-Medoids","K-Means"])
# select number of clusters
ks = sidebar.slider("Select number of clusters", min_value=2, max_value=10, value=2)
# select a dataframe to apply cluster on
data_type = sidebar.selectbox("Select a dataframe:", ["Train","Test1","Test2"])
st.markdown(f"## Dataframe selected {data_type}")
udf = dfs[data_type.lower()]
# if selected kmedoids, do respective operations
if calg == "K-Medoids":
st.markdown("### K-Medoids Clustering")
# if using PCA or not
use_pca = sidebar.radio("Use PCA?",["Yes","No"])
# if not using pca, do default clustering
if use_pca=="No":
st.markdown("### Not Using PCA")
inertias = []
for c in range(1,ks+1):
tdf = udf.copy()
X = tdf[features]
colors=['red','green','blue','magenta','black','yellow']
model = KMedoids(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=tdf[features[0]],y=tdf[features[1]],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(height=600, width=800, title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
# if using pca, use pca to reduce dimensionality and then do clustering
if use_pca=="Yes":
st.markdown("### Using PCA")
tdf=udf.copy()
X = udf[features]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)
feat = list(range(pca.n_components_))
PCA_components = pd.DataFrame(principalComponents, columns=list(range(len(feat))))
choosed_component = sidebar.multiselect("Choose Components",feat,default=[1,2])
choosed_component=[int(i) for i in choosed_component]
inertias = []
for c in range(1,ks+1):
X = PCA_components[choosed_component]
colors=['red','green','blue','magenta','black','yellow']
model = KMedoids(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
# if chosen KMeans, do respective operations
if calg == "K-Means":
st.markdown("### K-Means Clustering")
use_pca = sidebar.radio("Use PCA?",["Yes","No"])
if use_pca=="No":
st.markdown("### Not Using PCA")
inertias = []
for c in range(1,ks+1):
tdf = udf.copy()
X = tdf[features]
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=tdf[features[0]],y=tdf[features[1]],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(height=600, width=800, title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
if use_pca=="Yes":
st.markdown("### Using PCA")
tdf=udf.copy()
X = udf[features]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)
feat = list(range(pca.n_components_))
PCA_components = pd.DataFrame(principalComponents, columns=list(range(len(feat))))
choosed_component = sidebar.multiselect("Choose Components",feat,default=[1,2])
choosed_component=[int(i) for i in choosed_component]
inertias = []
for c in range(1,ks+1):
X = PCA_components[choosed_component]
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
Please refer to the comments for explanation of the code above. The web app should be something like below:
Now we will move on to the Regression part and implement it on our APP.
Regression
In this part, we will perform linear regression where we try to predict the occupancy based on other features. The metric will be calculated using model.score. The metric will be R2 Score.
from sklearn.linear_model import LinearRegression
model = LinearRegression()
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"]
dfs['btrain'] = pd.concat([train,test2])
xtest = dfs["test1"][features].to_numpy()
ytest = dfs["test1"]["Occupancy"]
for ddn,d in dfs.items():
if ddn!="test1":
print(ddn)
X = d[features].to_numpy().reshape(-1,len(features))
y = d["Occupancy"]
model.fit(X,y)
print(f"Model R2: {model.score(X,y)}")
print(f"Test R2: {model.score(xtest,ytest)}")
train
Model R2: 0.8580749633459134
Test R2: 0.8714317856126421
test2
Model R2: 0.8952863420051961
Test R2: 0.8658567155646273
btrain
Model R2: 0.8693410187120196
Test R2: 0.8649947193268359
Looking over the results above, btrain seems to have given a high R2 Score but train also have good test score.
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"]
for dn,d in dfs.items():
if ddn!="test1":
print(ddn)
X = d[features].to_numpy().reshape(-1,len(features))
y = d["Occupancy"]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
# Create a PCA instance: pca
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)
feat = range(pca.n_components_)
PCA_components = pd.DataFrame(principalComponents, columns=list(feat))
model=LinearRegression()
model.fit(PCA_components.to_numpy().reshape(-1,len(feat)),y)
print(f"Model R2: {model.score(PCA_components.to_numpy().reshape(-1,len(feat)),y)}")
#print(f"Test R2: {model.score(xtest,ytest)}")
btrain
Model R2: 0.8533646108336054
btrain
Model R2: 0.8786596706469451
btrain
Model R2: 0.6389469109358212
btrain
Model R2: 0.6305745433548783
It seems that our best model is from default Linear regression but still lets take PCA into the Streamlit app.
Taking Regression to Streamlit App
from sklearn.linear_model import LinearRegression, LogisticRegression, Ridge, Lasso, ElasticNet
if mode == "Regression":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"]
algorithm = sidebar.selectbox("Choose Algorithm",["Linear Regression","Ridge Regression","Lasso Regression","Elastic Net"])
st.markdown(f"### Chosen {algorithm}")
models = {"Linear Regression":LinearRegression(), "Ridge Regression":Ridge(), "Lasso Regression":Lasso(), "Elastic Net":ElasticNet()}
model = models[algorithm]
train_df = sidebar.selectbox("Choose Train Data",data_list)
test_df = sidebar.selectbox("Choose Test Data",[i for i in data_list if i != train_df])
selected = sidebar.multiselect("Choose Features",features,default=features)
xtrain = dfs[train_df][selected].to_numpy().reshape(-1,len(selected))
xtest = dfs[test_df][selected].to_numpy().reshape(-1,len(selected))
ytrain = dfs[train_df]["Occupancy"].to_numpy()
ytest = dfs[test_df]["Occupancy"].to_numpy()
model.fit(xtrain,ytrain)
st.markdown(f"Train R2 Score: {model.score(xtrain,ytrain)}")
st.markdown(f"Test R2 Score: {model.score(xtest,ytest)}")
- We have imported few regression algorithms from sklearn.
- We made a select box to select an algorithm.
- Made select box to choose train/test data.
- Made multi select box to choose features to use while making a model.
- Then we trained a model using selected data, selected feature and selected algorithm.
- Printed the accuracy also.

Adding few more lines of codes to show coefficient and take user input for a prediction:
if sidebar.checkbox("Show Coefficients"):
st.markdown("#### Showing Coefficents and Intercept")
st.write(f"Coeffs: {model.coef_}")
st.write(f"Intercept: {model.intercept_}")
if sidebar.checkbox("Show Prediction"):
st.markdown("#### Showing Prediction")
input_values = [float((st.number_input(t))) for t in selected]
prediction = model.predict([input_values])
st.write(f"Predicted {prediction}")
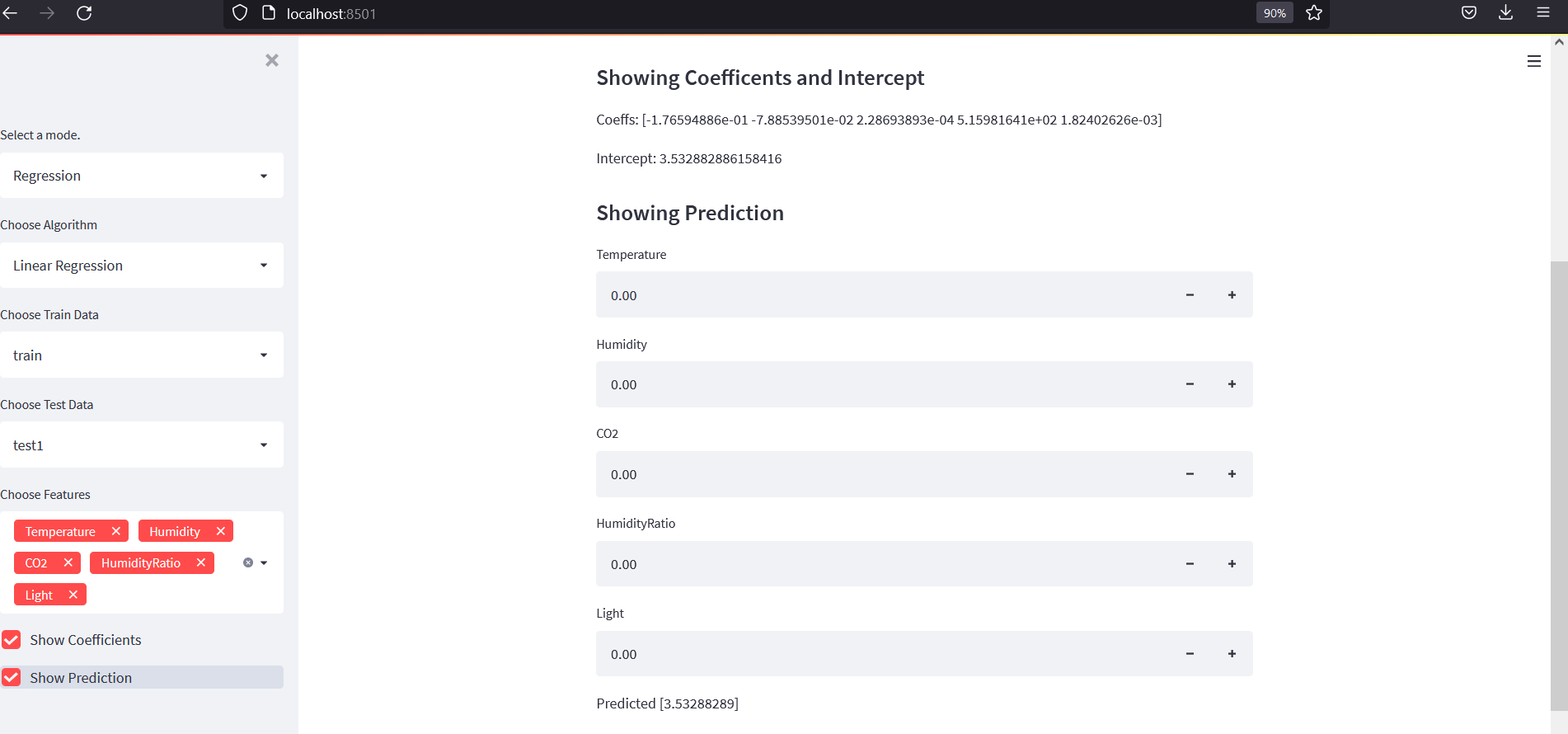
All Codes
Below is the codes that we wrote upto now.
import streamlit as st
import numpy as np
import pandas as pd
import cufflinks
from plotly.subplots import make_subplots
import plotly.graph_objects as go
from sklearn_extra.cluster import KMedoids
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LinearRegression, LogisticRegression, Ridge, Lasso, ElasticNet
hline="--"*40
@st.cache
def get_data():
train = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatraining.txt")
train["date"]=pd.to_datetime(train.date)
test1 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest.txt")
test1["date"]=pd.to_datetime(test1.date)
test2 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest2.txt")
test2["date"]=pd.to_datetime(test2.date)
dfs = {"train":train,"test1":test1,"test2":test2}
return dfs
dfs = get_data()
data_list = list(dfs.keys())
sidebar = st.sidebar
# select modes, EDA, Clustering, Regression and Classification
mode = sidebar.selectbox("Select a mode.",options=["EDA", "Clustering", "Regression", "Classification"])
st.markdown(f"### {mode} Mode Selected")
st.markdown(hline)
# If selected EDA, show EDA related plots
if mode=="EDA":
# if selected show the data
show_data = sidebar.checkbox("Show data")
if show_data:
# if selected, show train data
if sidebar.checkbox("Show Train data"):
st.markdown("### Train Data")
st.dataframe(dfs["train"])
# if selected, show test1 data
if sidebar.checkbox("Show Test1 data"):
st.markdown("### Test1 Data")
st.dataframe(dfs["test1"])
# if selected, show test2 data
if sidebar.checkbox("Show Test2 data"):
st.markdown("### Test2 Data")
st.dataframe(dfs["test2"])
# if selected, show the comparision data
show_comparison = sidebar.checkbox("Show comparison")
if show_comparison:
# make a multiselect to select the columns to compare
selected = sidebar.multiselect("Select Columns ", [d for d in dfs["train"].columns if d not in ["date"]])
titles=list(dfs.keys())
train = dfs["train"]
test1 = dfs["test1"]
test2 = dfs["test2"]
if selected:
st.markdown(f"### Selected Columns: {', '.join(selected)}")
for c in selected:
fig = make_subplots(rows=2,cols=3, subplot_titles=titles, )
fig.add_trace(go.Box(y=train[c].tolist(), name=titles[0]), row=1, col=1)
fig.add_trace(go.Box(y=test1[c].tolist(), name = titles[1]), row=1, col=2)
fig.add_trace(go.Box(y=test2[c].tolist(), name = titles[2]), row=1, col=3)
fig.add_trace(go.Histogram(y=train[c].tolist(), name=titles[0]), row=2, col=1)
fig.add_trace(go.Histogram(y=test1[c].tolist(), name = titles[1]), row=2, col=2)
fig.add_trace(go.Histogram(y=test2[c].tolist(), name = titles[2]), row=2, col=3)
fig.update_layout(height=600, width=800, title_text=f"Box and Distribution of {c}")
st.plotly_chart(fig)
# if selected show correlation
show_corr = sidebar.checkbox("Show Correlation")
if show_corr:
st.markdown("### Correlation")
fig = make_subplots(rows=1,cols=3, subplot_titles=titles)
fig.add_trace(go.Heatmap(z=train.corr(), y=train.corr().columns,x=train.corr().index, name=titles[0]), row=1, col=1)
fig.add_trace(go.Heatmap(z=test1.corr(), x=train.corr().index, name = titles[1]), row=1, col=2)
fig.add_trace(go.Heatmap(z=test2.corr(), x=train.corr().index, name = titles[2]), row=1, col=3)
st.plotly_chart(fig)
if mode=="Clustering":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"][:-2]
# select a clustering algorithm
calg = sidebar.selectbox("Select a clustering algorithm", ["K-Medoids","K-Means"])
# select number of clusters
ks = sidebar.slider("Select number of clusters", min_value=2, max_value=10, value=2)
# select a dataframe to apply cluster on
data_type = sidebar.selectbox("Select a dataframe:", ["Train","Test1","Test2"])
st.markdown(f"## Dataframe selected {data_type}")
udf = dfs[data_type.lower()]
# if selected kmedoids, do respective operations
if calg == "K-Medoids":
st.markdown("### K-Medoids Clustering")
# if using PCA or not
use_pca = sidebar.radio("Use PCA?",["Yes","No"])
# if not using pca, do default clustering
if use_pca=="No":
st.markdown("### Not Using PCA")
inertias = []
for c in range(1,ks+1):
tdf = udf.copy()
X = tdf[features]
colors=['red','green','blue','magenta','black','yellow']
model = KMedoids(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=tdf[features[0]],y=tdf[features[1]],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(height=600, width=800, title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
# if using pca, use pca to reduce dimensionality and then do clustering
if use_pca=="Yes":
st.markdown("### Using PCA")
tdf=udf.copy()
X = udf[features]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)
feat = list(range(pca.n_components_))
PCA_components = pd.DataFrame(principalComponents, columns=list(range(len(feat))))
choosed_component = sidebar.multiselect("Choose Components",feat,default=[1,2])
choosed_component=[int(i) for i in choosed_component]
inertias = []
for c in range(1,ks+1):
X = PCA_components[choosed_component]
colors=['red','green','blue','magenta','black','yellow']
model = KMedoids(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
# if chosen KMeans, do respective operations
if calg == "K-Means":
st.markdown("### K-Means Clustering")
use_pca = sidebar.radio("Use PCA?",["Yes","No"])
if use_pca=="No":
st.markdown("### Not Using PCA")
inertias = []
for c in range(1,ks+1):
tdf = udf.copy()
X = tdf[features]
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=tdf[features[0]],y=tdf[features[1]],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(height=600, width=800, title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
if use_pca=="Yes":
st.markdown("### Using PCA")
tdf=udf.copy()
X = udf[features]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)
feat = list(range(pca.n_components_))
PCA_components = pd.DataFrame(principalComponents, columns=list(range(len(feat))))
choosed_component = sidebar.multiselect("Choose Components",feat,default=[1,2])
choosed_component=[int(i) for i in choosed_component]
inertias = []
for c in range(1,ks+1):
X = PCA_components[choosed_component]
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
if mode == "Regression":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"]
algorithm = sidebar.selectbox("Choose Algorithm",["Linear Regression","Ridge Regression","Lasso Regression","Elastic Net"])
st.markdown(f"### Chosen {algorithm}")
models = {"Linear Regression":LinearRegression(), "Ridge Regression":Ridge(), "Lasso Regression":Lasso(), "Elastic Net":ElasticNet()}
model = models[algorithm]
train_df = sidebar.selectbox("Choose Train Data",data_list)
test_df = sidebar.selectbox("Choose Test Data",[i for i in data_list if i != train_df])
selected = sidebar.multiselect("Choose Features",features,default=features)
xtrain = dfs[train_df][selected].to_numpy().reshape(-1,len(selected))
xtest = dfs[test_df][selected].to_numpy().reshape(-1,len(selected))
ytrain = dfs[train_df]["Occupancy"].to_numpy()
ytest = dfs[test_df]["Occupancy"].to_numpy()
model.fit(xtrain,ytrain)
st.markdown(f"Train R2 Score: {model.score(xtrain,ytrain)}")
st.markdown(f"Test R2 Score: {model.score(xtest,ytest)}")
if sidebar.checkbox("Show Coefficients"):
st.markdown("#### Showing Coefficents and Intercept")
st.write(f"Coeffs: {model.coef_}")
st.write(f"Intercept: {model.intercept_}")
if sidebar.checkbox("Show Prediction"):
st.markdown("#### Showing Prediction")
input_values = [float((st.number_input(t))) for t in selected]
prediction = model.predict([input_values])
st.write(f"Predicted {prediction}")
Classification
Taking Classification to Streamlit App
Until now we have created EDA, Clustering and Regression modes now is the time for us to create a classification models. We have covered a most of the redundant part of try out on above sections but in this one, we will jump right into the implementation part.
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import confusion_matrix, f1_score, accuracy_score
if mode == "Classification":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"]
algorithm = sidebar.selectbox("Choose Algorithm",["Logistic Regression","KNN","Decision Tree","Random Forest", "Ada Boost"])
st.markdown(f"### Chosen {algorithm}")
models = {"Logistic Regression":LogisticRegression(), "KNN":KNeighborsClassifier(), "Decision Tree":DecisionTreeClassifier(), "Random Forest":RandomForestClassifier(), "Ada Boost":AdaBoostClassifier()}
model = models[algorithm]
train_df = sidebar.selectbox("Choose Train Data",data_list)
test_df = sidebar.selectbox("Choose Test Data",[i for i in data_list if i != train_df])
selected = sidebar.multiselect("Choose Features",features,default=features)
xtrain = dfs[train_df][selected].to_numpy().reshape(-1,len(selected))
xtest = dfs[test_df][selected].to_numpy().reshape(-1,len(selected))
ytrain = dfs[train_df]["Occupancy"].to_numpy()
ytest = dfs[test_df]["Occupancy"].to_numpy()
model.fit(xtrain,ytrain)
st.markdown(f"##### R2 Score: Train = {model.score(xtrain,ytrain) : .2f}, Test = {model.score(xtest,ytest) : .2f}")
train_pred = model.predict(xtrain)
test_pred = model.predict(xtest)
cm_train = confusion_matrix(ytrain,train_pred,labels=[0,1])
cm_test = confusion_matrix(ytest,test_pred,labels=[0,1])
train_f1 = f1_score(ytrain,train_pred,average="macro")
test_f1 = f1_score(ytest,test_pred,average="macro")
train_acc = accuracy_score(ytrain,train_pred)
test_acc = accuracy_score(ytest,test_pred)
st.markdown(f"##### F1 Score: Train = {train_f1 : .2f}, Test = {test_f1 : .2f}")
st.markdown(f"##### Accuracy Score: Train = {train_acc : .2f}, Test = {test_acc : .2f}")
fig = make_subplots(rows=1,cols=2, subplot_titles=["Train","Test"])
labels = ["Vacant","Occupied"]
fig1 = go.Heatmap(z=cm_train, y=labels, x=labels, name="Train")
fig2 = go.Heatmap(z=cm_test, y=labels, x=labels, name="Test")
fig.add_trace(fig1, row=1, col=1)
fig.add_trace(fig2, row=1, col=2)
fig.update_layout({"title":"Confusion Matrix","xaxis": {"title": "Predicted value"},
"yaxis": {"title": "Real value"}})
st.plotly_chart(fig)
if sidebar.checkbox("Show Coefficients"):
st.markdown("#### Showing Coefficents and Intercept")
try:
st.write(f"Coeffs: {model.coef_}")
st.write(f"Intercept: {model.intercept_}")
except:
st.write("Coeffs: Not Available")
if sidebar.checkbox("Show Prediction"):
st.markdown("#### Showing Prediction")
input_values = [float((st.number_input(t))) for t in selected]
prediction = model.predict([input_values])
st.write(f"Predicted {prediction}")
What we are doing in above code is:
- Select the classification algorithm and make its class.
- Choose features and then prepare data using that features.
- Train a model and show train/test metrics.
- Show confusion matrix.
If everything is fine, then our app should look like below:
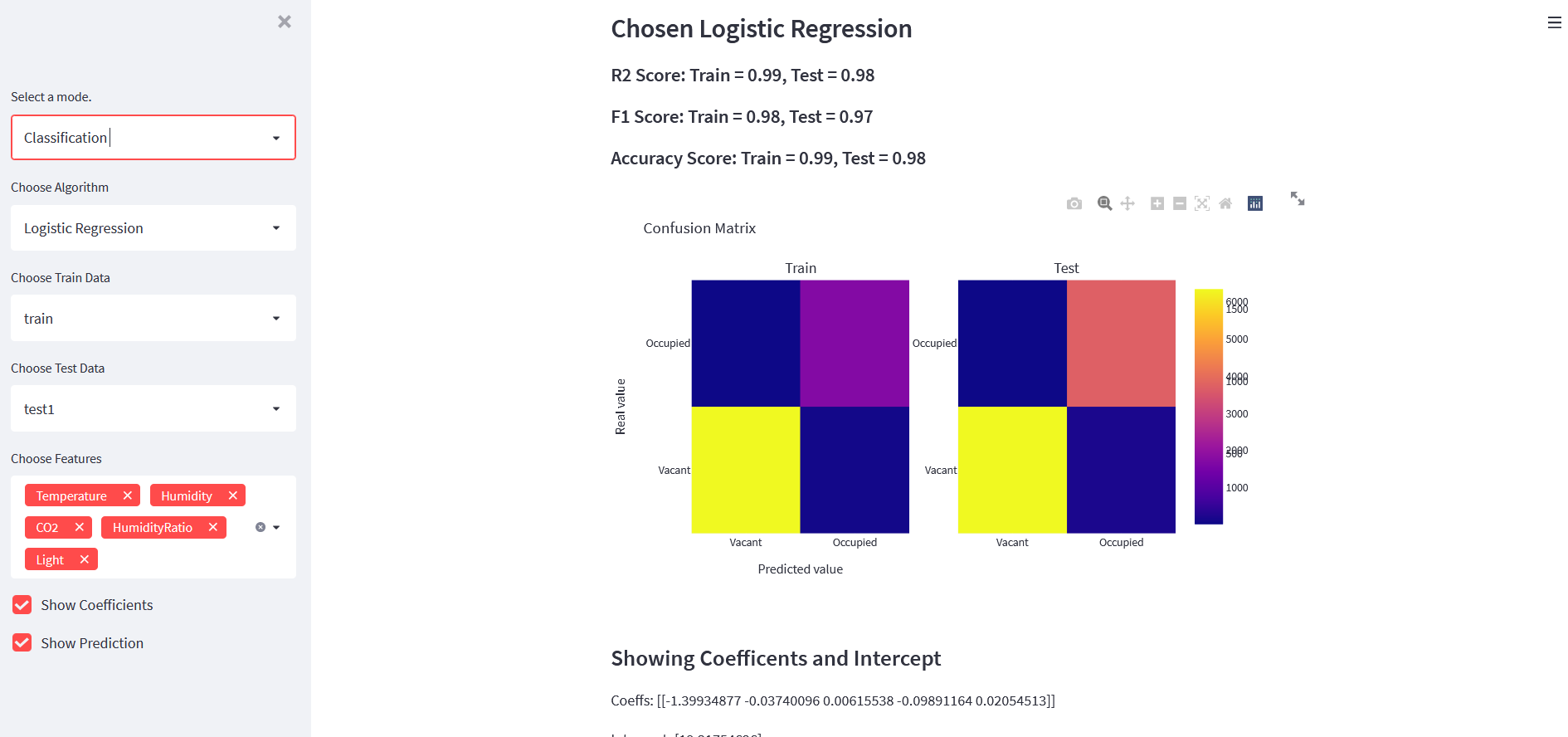
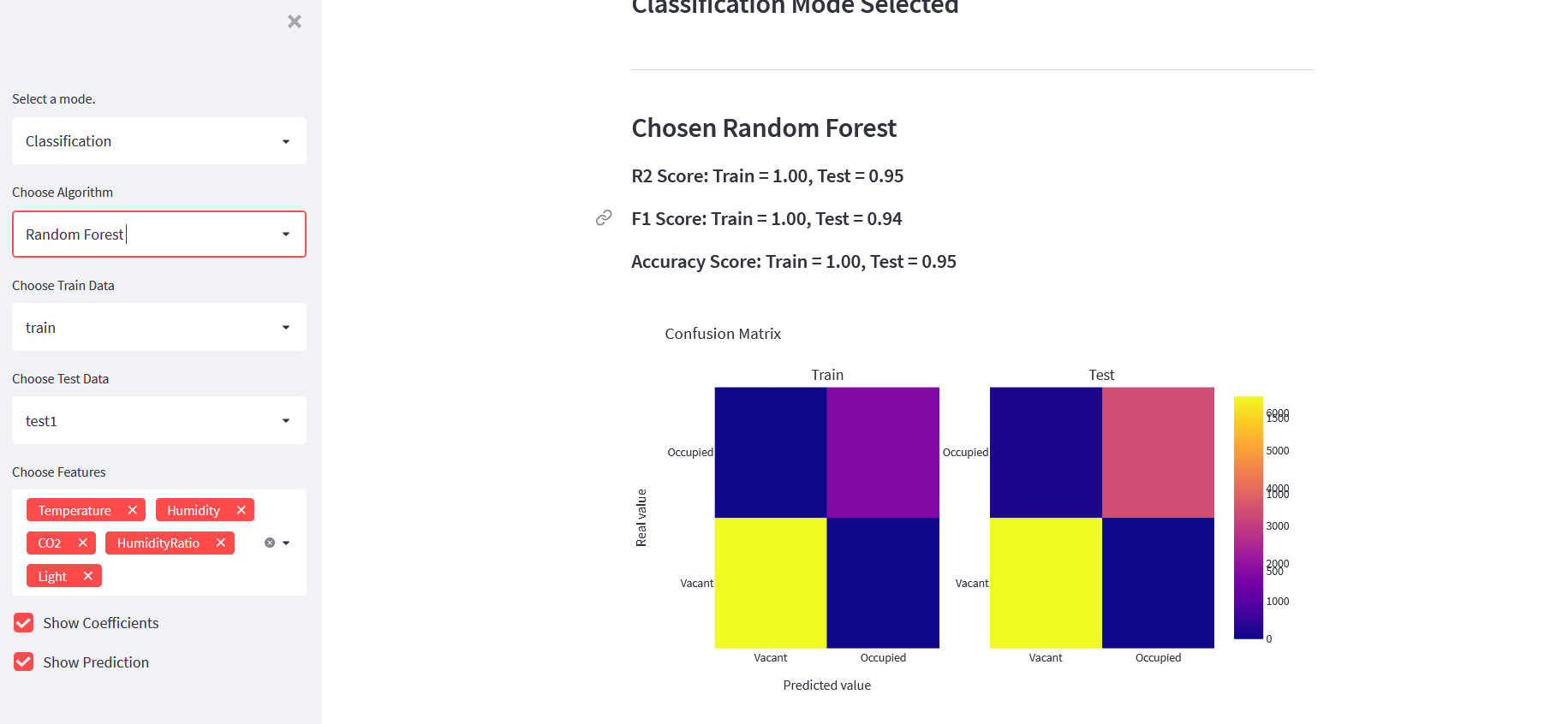
All Codes
from sklearn.metrics import plot_confusion_matrix
import streamlit as st
import numpy as np
import pandas as pd
import cufflinks
from plotly.subplots import make_subplots
import plotly.graph_objects as go
from sklearn_extra.cluster import KMedoids
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LinearRegression, LogisticRegression, Ridge, Lasso, ElasticNet
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import confusion_matrix, f1_score, accuracy_score
hline="--"*40
@st.cache
def get_data():
train = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatraining.txt")
train["date"]=pd.to_datetime(train.date)
test1 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest.txt")
test1["date"]=pd.to_datetime(test1.date)
test2 = pd.read_csv("https://github.com/LuisM78/Occupancy-detection-data/raw/master/datatest2.txt")
test2["date"]=pd.to_datetime(test2.date)
dfs = {"train":train,"test1":test1,"test2":test2}
return dfs
dfs = get_data()
data_list = list(dfs.keys())
sidebar = st.sidebar
# select modes, EDA, Clustering, Regression and Classification
mode = sidebar.selectbox("Select a mode.",options=["EDA", "Clustering", "Regression", "Classification"])
st.markdown(f"### {mode} Mode Selected")
st.markdown(hline)
# If selected EDA, show EDA related plots
if mode=="EDA":
# if selected show the data
show_data = sidebar.checkbox("Show data")
if show_data:
# if selected, show train data
if sidebar.checkbox("Show Train data"):
st.markdown("### Train Data")
st.dataframe(dfs["train"])
# if selected, show test1 data
if sidebar.checkbox("Show Test1 data"):
st.markdown("### Test1 Data")
st.dataframe(dfs["test1"])
# if selected, show test2 data
if sidebar.checkbox("Show Test2 data"):
st.markdown("### Test2 Data")
st.dataframe(dfs["test2"])
# if selected, show the comparision data
show_comparison = sidebar.checkbox("Show comparison")
if show_comparison:
# make a multiselect to select the columns to compare
selected = sidebar.multiselect("Select Columns ", [d for d in dfs["train"].columns if d not in ["date"]])
titles=list(dfs.keys())
train = dfs["train"]
test1 = dfs["test1"]
test2 = dfs["test2"]
if selected:
st.markdown(f"### Selected Columns: {', '.join(selected)}")
for c in selected:
fig = make_subplots(rows=2,cols=3, subplot_titles=titles, )
fig.add_trace(go.Box(y=train[c].tolist(), name=titles[0]), row=1, col=1)
fig.add_trace(go.Box(y=test1[c].tolist(), name = titles[1]), row=1, col=2)
fig.add_trace(go.Box(y=test2[c].tolist(), name = titles[2]), row=1, col=3)
fig.add_trace(go.Histogram(y=train[c].tolist(), name=titles[0]), row=2, col=1)
fig.add_trace(go.Histogram(y=test1[c].tolist(), name = titles[1]), row=2, col=2)
fig.add_trace(go.Histogram(y=test2[c].tolist(), name = titles[2]), row=2, col=3)
fig.update_layout(height=600, width=800, title_text=f"Box and Distribution of {c}")
st.plotly_chart(fig)
# if selected show correlation
show_corr = sidebar.checkbox("Show Correlation")
if show_corr:
st.markdown("### Correlation")
fig = make_subplots(rows=1,cols=3, subplot_titles=titles)
fig.add_trace(go.Heatmap(z=train.corr(), y=train.corr().columns,x=train.corr().index, name=titles[0]), row=1, col=1)
fig.add_trace(go.Heatmap(z=test1.corr(), x=train.corr().index, name = titles[1]), row=1, col=2)
fig.add_trace(go.Heatmap(z=test2.corr(), x=train.corr().index, name = titles[2]), row=1, col=3)
st.plotly_chart(fig)
if mode=="Clustering":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"][:-2]
# select a clustering algorithm
calg = sidebar.selectbox("Select a clustering algorithm", ["K-Medoids","K-Means"])
# select number of clusters
ks = sidebar.slider("Select number of clusters", min_value=2, max_value=10, value=2)
# select a dataframe to apply cluster on
data_type = sidebar.selectbox("Select a dataframe:", ["Train","Test1","Test2"])
st.markdown(f"## Dataframe selected {data_type}")
udf = dfs[data_type.lower()]
# if selected kmedoids, do respective operations
if calg == "K-Medoids":
st.markdown("### K-Medoids Clustering")
# if using PCA or not
use_pca = sidebar.radio("Use PCA?",["Yes","No"])
# if not using pca, do default clustering
if use_pca=="No":
st.markdown("### Not Using PCA")
inertias = []
for c in range(1,ks+1):
tdf = udf.copy()
X = tdf[features]
colors=['red','green','blue','magenta','black','yellow']
model = KMedoids(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=tdf[features[0]],y=tdf[features[1]],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(height=600, width=800, title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
# if using pca, use pca to reduce dimensionality and then do clustering
if use_pca=="Yes":
st.markdown("### Using PCA")
tdf=udf.copy()
X = udf[features]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)
feat = list(range(pca.n_components_))
PCA_components = pd.DataFrame(principalComponents, columns=list(range(len(feat))))
choosed_component = sidebar.multiselect("Choose Components",feat,default=[1,2])
choosed_component=[int(i) for i in choosed_component]
inertias = []
for c in range(1,ks+1):
X = PCA_components[choosed_component]
colors=['red','green','blue','magenta','black','yellow']
model = KMedoids(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
# if chosen KMeans, do respective operations
if calg == "K-Means":
st.markdown("### K-Means Clustering")
use_pca = sidebar.radio("Use PCA?",["Yes","No"])
if use_pca=="No":
st.markdown("### Not Using PCA")
inertias = []
for c in range(1,ks+1):
tdf = udf.copy()
X = tdf[features]
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=tdf[features[0]],y=tdf[features[1]],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(height=600, width=800, title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
if use_pca=="Yes":
st.markdown("### Using PCA")
tdf=udf.copy()
X = udf[features]
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
pca = PCA(n_components=3)
principalComponents = pca.fit_transform(X_scaled)
feat = list(range(pca.n_components_))
PCA_components = pd.DataFrame(principalComponents, columns=list(range(len(feat))))
choosed_component = sidebar.multiselect("Choose Components",feat,default=[1,2])
choosed_component=[int(i) for i in choosed_component]
inertias = []
for c in range(1,ks+1):
X = PCA_components[choosed_component]
colors=['red','green','blue','magenta','black','yellow']
model = KMeans(n_clusters=c)
model.fit(X)
y_kmeans = model.predict(X)
tdf["cluster"] = y_kmeans
inertias.append((c,model.inertia_))
trace0 = go.Scatter(x=X[1],y=X[2],mode='markers', marker=dict(
color=tdf.cluster.apply(lambda x: colors[x]),
colorscale='Viridis',
showscale=True
),name="Cluster Points")
trace1 = go.Scatter(x=model.cluster_centers_[:, 0], y=model.cluster_centers_[:, 1],mode='markers', marker=dict(
color=colors,
size=20,
showscale=True
),name="Cluster Mean")
data7 = go.Data([trace0, trace1])
fig = go.Figure(data=data7)
fig.update_layout(title=f"Cluster Size {c}")
st.plotly_chart(fig)
inertias=np.array(inertias).reshape(-1,2)
performance = go.Scatter(x=inertias[:,0], y=inertias[:,1])
layout = go.Layout(
title="Cluster Number vs Inertia",
xaxis=dict(
title="Ks"
),
yaxis=dict(
title="Inertia"
) )
fig=go.Figure(data=go.Data([performance]))
fig.update_layout(layout)
st.plotly_chart(fig)
if mode == "Regression":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"]
algorithm = sidebar.selectbox("Choose Algorithm",["Linear Regression","Ridge Regression","Lasso Regression","Elastic Net"])
st.markdown(f"### Chosen {algorithm}")
models = {"Linear Regression":LinearRegression(), "Ridge Regression":Ridge(), "Lasso Regression":Lasso(), "Elastic Net":ElasticNet()}
model = models[algorithm]
train_df = sidebar.selectbox("Choose Train Data",data_list)
test_df = sidebar.selectbox("Choose Test Data",[i for i in data_list if i != train_df])
selected = sidebar.multiselect("Choose Features",features,default=features)
xtrain = dfs[train_df][selected].to_numpy().reshape(-1,len(selected))
xtest = dfs[test_df][selected].to_numpy().reshape(-1,len(selected))
ytrain = dfs[train_df]["Occupancy"].to_numpy()
ytest = dfs[test_df]["Occupancy"].to_numpy()
model.fit(xtrain,ytrain)
st.markdown(f"Train R2 Score: {model.score(xtrain,ytrain) : .2f}")
st.markdown(f"Test R2 Score: {model.score(xtest,ytest) : .2f}")
if sidebar.checkbox("Show Coefficients"):
st.markdown("#### Showing Coefficents and Intercept")
st.write(f"Coeffs: {model.coef_}")
st.write(f"Intercept: {model.intercept_}")
if sidebar.checkbox("Show Prediction"):
st.markdown("#### Showing Prediction")
input_values = [float((st.number_input(t))) for t in selected]
prediction = model.predict([input_values])
st.write(f"Predicted {prediction}")
if mode == "Classification":
features = ["Temperature", "Humidity", "CO2", "HumidityRatio","Light"]
algorithm = sidebar.selectbox("Choose Algorithm",["Logistic Regression","KNN","Decision Tree","Random Forest", "Ada Boost"])
st.markdown(f"### Chosen {algorithm}")
models = {"Logistic Regression":LogisticRegression(), "KNN":KNeighborsClassifier(), "Decision Tree":DecisionTreeClassifier(), "Random Forest":RandomForestClassifier(), "Ada Boost":AdaBoostClassifier()}
model = models[algorithm]
train_df = sidebar.selectbox("Choose Train Data",data_list)
test_df = sidebar.selectbox("Choose Test Data",[i for i in data_list if i != train_df])
selected = sidebar.multiselect("Choose Features",features,default=features)
xtrain = dfs[train_df][selected].to_numpy().reshape(-1,len(selected))
xtest = dfs[test_df][selected].to_numpy().reshape(-1,len(selected))
ytrain = dfs[train_df]["Occupancy"].to_numpy()
ytest = dfs[test_df]["Occupancy"].to_numpy()
model.fit(xtrain,ytrain)
st.markdown(f"##### R2 Score: Train = {model.score(xtrain,ytrain) : .2f}, Test = {model.score(xtest,ytest) : .2f}")
train_pred = model.predict(xtrain)
test_pred = model.predict(xtest)
cm_train = confusion_matrix(ytrain,train_pred,labels=[0,1])
cm_test = confusion_matrix(ytest,test_pred,labels=[0,1])
train_f1 = f1_score(ytrain,train_pred,average="macro")
test_f1 = f1_score(ytest,test_pred,average="macro")
train_acc = accuracy_score(ytrain,train_pred)
test_acc = accuracy_score(ytest,test_pred)
st.markdown(f"##### F1 Score: Train = {train_f1 : .2f}, Test = {test_f1 : .2f}")
st.markdown(f"##### Accuracy Score: Train = {train_acc : .2f}, Test = {test_acc : .2f}")
fig = make_subplots(rows=1,cols=2, subplot_titles=["Train","Test"])
labels = ["Vacant","Occupied"]
fig1 = go.Heatmap(z=cm_train, y=labels, x=labels, name="Train")
fig2 = go.Heatmap(z=cm_test, y=labels, x=labels, name="Test")
fig.add_trace(fig1, row=1, col=1)
fig.add_trace(fig2, row=1, col=2)
fig.update_layout({"title":"Confusion Matrix","xaxis": {"title": "Predicted value"},
"yaxis": {"title": "Real value"}})
st.plotly_chart(fig)
if sidebar.checkbox("Show Coefficients"):
st.markdown("#### Showing Coefficents and Intercept")
try:
st.write(f"Coeffs: {model.coef_}")
st.write(f"Intercept: {model.intercept_}")
except:
st.write("Coeffs: Not Available")
if sidebar.checkbox("Show Prediction"):
st.markdown("#### Showing Prediction")
input_values = [float((st.number_input(t))) for t in selected]
prediction = model.predict([input_values])
st.write(f"Predicted {prediction}")
Add Inference Mode
For this purpose, we will need to save a model during Regression or Classification phase and then upload it on inference to test it. What we will do is,
- Accept a model file upload and read number of features in it.
- Create a form where we will have number of input fields equal to number of features to accept input vlaues.
- Create a submit button to pass those input values to loaded model and then print the result.
if mode in ["Regression", "Classification"]:
filename=sidebar.text_input("Enter File Name",value="model.sav")
save = sidebar.button("Save Model")
if save and model:
pickle.dump(model, open(filename, 'wb'))
sidebar.markdown("Saved Model!")
if mode =="Inference":
model=None
if "temp_model.csv" not in os.listdir():
file = sidebar.file_uploader("Upload Model File", accept_multiple_files=False)
if file:
model = pickle.load(file)
pickle.dump(model, open("temp_model.sav", 'wb'))
st.markdown("Model Loaded.")
if model:
st.markdown(f"Loaded {type(model).__name__} Model!")
nfeatures="Not Known"
try:
nfeatures=model.n_features_
except:
nfeatures = model.coef_.shape[-1]
st.markdown(f"Number of features: {nfeatures}")
if nfeatures!="Not Known":
form1 = st.form(key='my_form')
st.markdown("#### Showing Prediction")
input_values = [float((form1.number_input(f"Feature {t} Value"))) for t in range(nfeatures)]
submit= form1.form_submit_button("Predict")
prediction=None
if submit:
prediction = model.predict([input_values])
st.markdown(f"Input Values: {input_values}")
st.write(f"Predicted {prediction}")
Now we should be able to see a text field to enter a file name and then save button on Regression and Classification Mode.
Lets save some models and load them on Inference mode and try to predict from it. And our output should look like below:
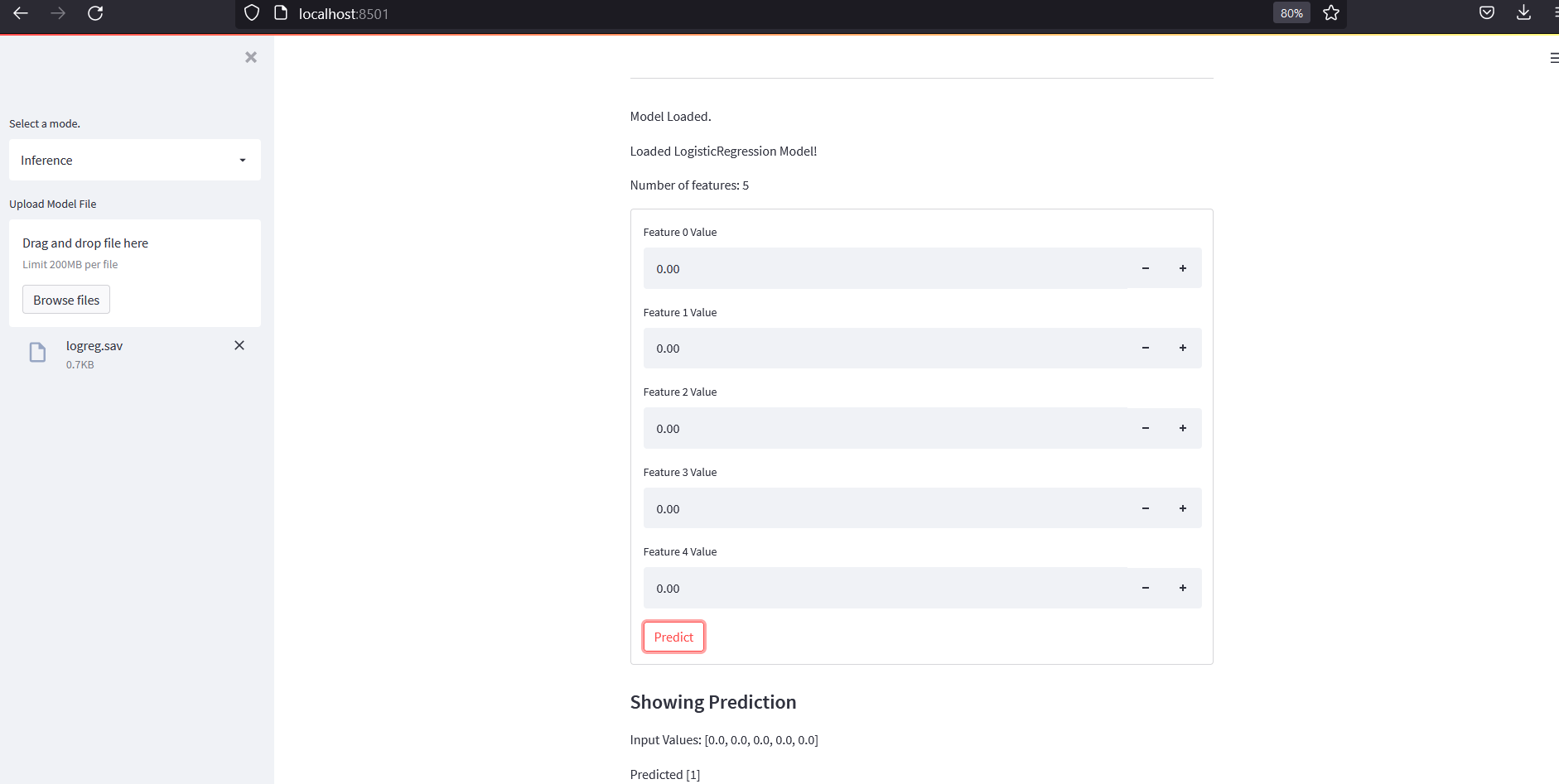
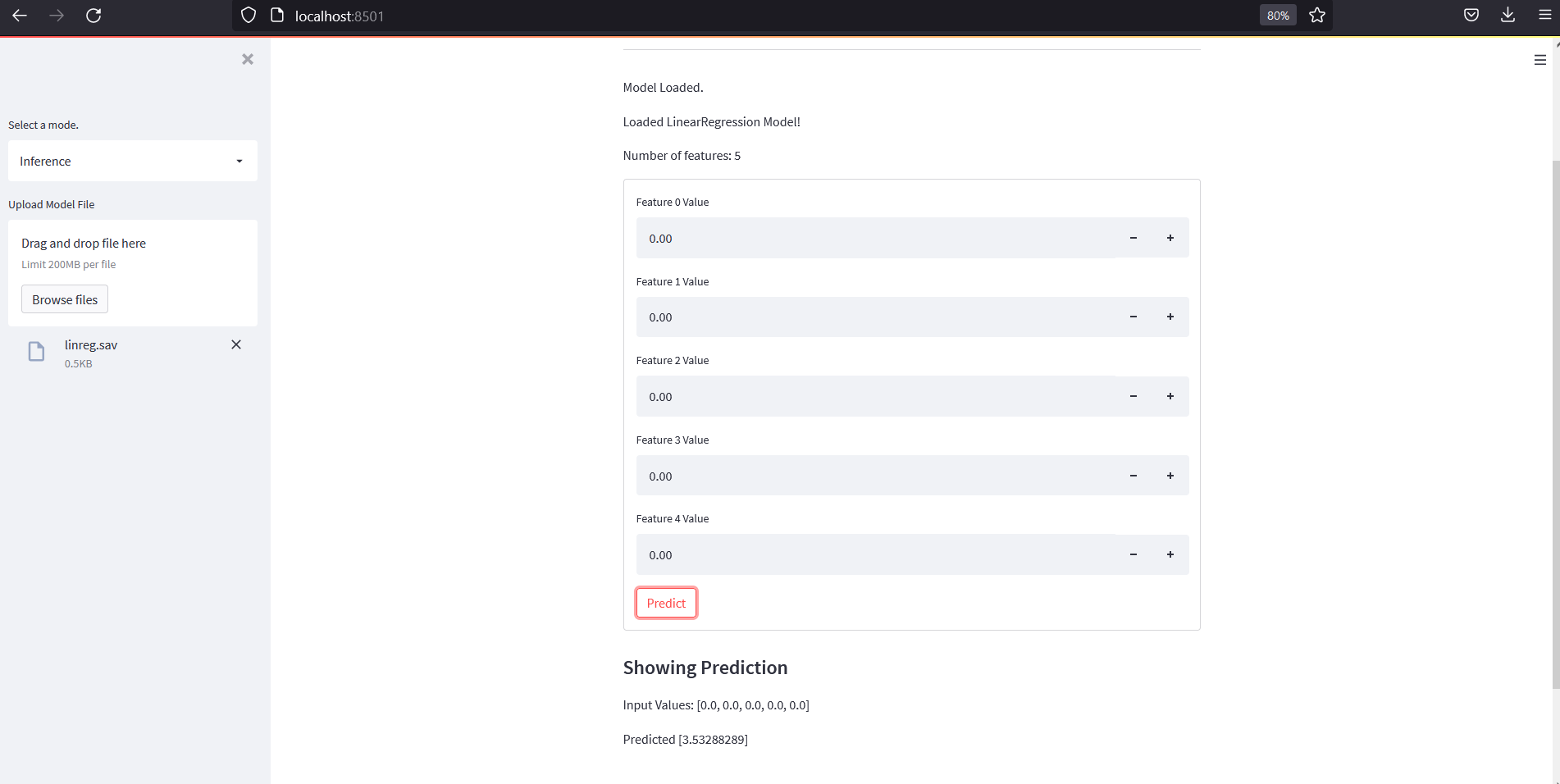
Conclusion
It was a long ride from past few projects, where we done some EDA, then explored possible ML Models to predict, cluster data and in this part, we tried to do all those things from the web app and also we can now try to predict from the webapp by inserting some values into the form.



Comments